Jupyter
Jupyter Notebook and JupyterLab are available through our cluster Web Portals. Information on accessing the web portal is available on Access the Web Portal documentation page. From the Web Portal, you are able to launch interactive apps such as Jupyter.
Launch Jupyter
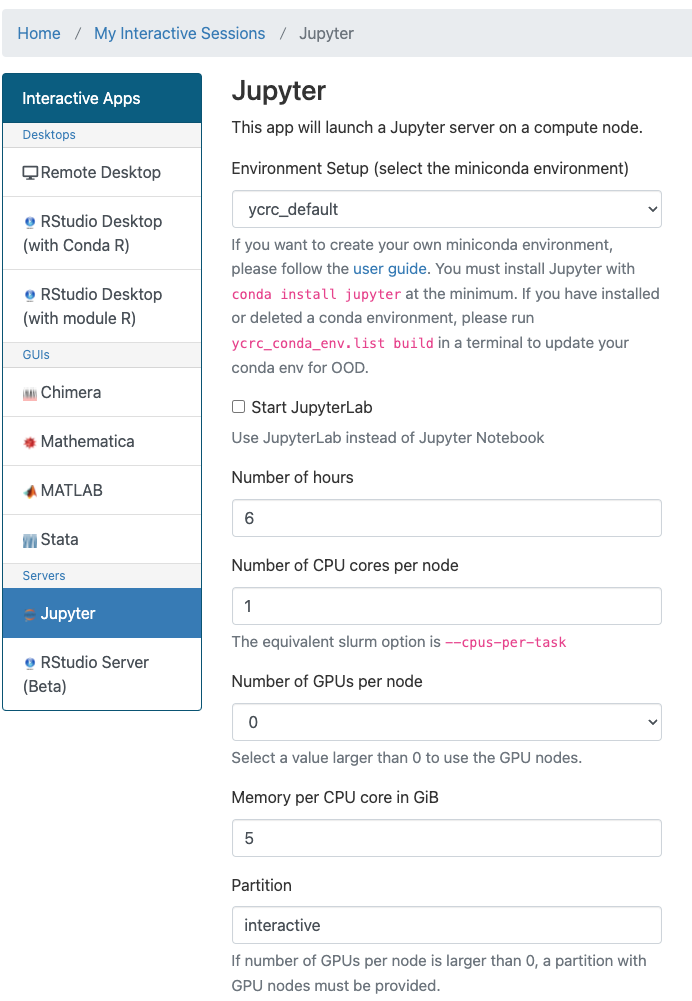
To get started, connect to one of cluster Web Portals and choose Jupyter from the Interactive Apps menu or the dashboard and then follow the instructions for launching an interactive app.
You can use the ycrc_default environment or chose one of your own from the drop-down menu.
After specifying the required resources (number of CPUs/GPUs, amount of RAM, etc.) and time limit, you can submit the job.
When it launches you can open the standard Jupyter interface where you can start working with notebooks.
Tip
If you have installed and want to use Jupyter Lab instead of Jupyter Notebook, check the Start JupyterLab checkbox.
If there is a specific workflow which OOD does not satisfy, let us know and we can help.
Root Directory
The Jupyter root directory is set to your Home when started. Project and Scratch can be accessed via their respective symlinks in Home. If you want to access a directory that cannot be accessed through your home directory, for example a purchased storage allocation, you need to create a symlink to that directory in your home directory.
Conda Environments
Make sure that you chose the correct Conda environment for your job from the drop-down menu.
ycrc_default
The ycrc_default conda environment will be automatically built when you select it for the first time from the Jupyter app.
Set Up an Environment
We recommend you use miniconda to manage your Jupyter environments. You can create Conda environments from the Web Portal terminal interface or from a terminal-based login to the clusters.
Warning
Please do not use the '--prefix' option when creating your own jupyter/conda environment for OOD; you must use '-n/--name'. This guarantees that your conda environment is located your home directory under '.conda/envs', which is required by our current OnDemand setup. (note, however, that this '.conda' folder, or 'envs' within it, can be a symlink pointing elsewhere)
For example, if you want to create an environment with many commonly used scientific computing Python packages you would run:
###request a compute node on the devel partition for 2 hours with 4 cpus and 15 GB of RAM
salloc --partition=devel --mem=15G --time=2:00:00 --cpus-per-task=2
module load miniconda
# To create a Jupyter/OOD compatible conda environment from scratch:
conda create -y -n notebook_env python notebook numpy pandas matplotlib
# Alternatively, to make an existing environment compatible with Jupyter OOD:
conda activate notebook_env
conda install notebook
# To add your environment to the OOD Jupyter miniconda menu:
ycrc_conda_env.sh update
Note that your conda environment must include notebook in its installed packages, using either the original conda create command or subsequently, with conda activate <...>; conda install notebook as above. Otherwise, the conda environment will fail inside of the OpenOndemand Jupyter instance.
The ycrc_conda_env.sh update command above is also important. Without it, your conda environment list on the Jupyter form will not update automatically. To update the list you must run this command.
Command-Line Execution of Jupyter Notebooks
Many scientific workflows start as interactive Jupyter notebooks, and our Web Portal has dramatically simplified deploying these notebooks on cluster resources. However, the step from running notebooks interactively to running jobs as a batch script can be challenging and is often a barrier to migrating to using sbatch to run workflows non-interactively.
To help solve this problem, there are a handful of utilities that can execute a notebook as if you were manually hitting "shift-Enter" for each cell. Of note is Papermill which provides a powerful set of tools to bridge between interactive and batch-mode computing.
To get started, install papermill into your conda environments:
module load miniconda
conda activate my_env
conda install papermill
Then you can simply evaluate a notebook, preserving figures and output inside the notebook, like this:
papermill /path/to/notebook.ipynb /path/to/output.ipynb
This can be run inside a batch job that might look like this:
#!/bin/bash
#SBATCH -p day
#SBATCH -c 1
#SBATCH -t 6:00:00
module reset
module load miniconda
conda activate my_env
papermill /path/to/notebook.ipynb /path/to/output.ipynb
Variables can also be parameterized and passed in as command-line options so that you can run multiple copies simultaneously with different input variables. For more information see the Papermill docs pages.
Troubleshoot
Jupyter cannot be started properly
- If you are trying to launch
jupyter-notebook, make sure it is available in your jupyter conda environment:(ycrc_default)[pl543@grace1 ~]$ which jupyter-notebook /gpfs/gibbs/project/support/pl543/conda_envs/ycrc_default/bin/jupyter-notebook - If you are trying to launch
jupyter-lab, make sure it is available in your jupyter conda environment:(ycrc_default)[pl543@grace1 ~]$ which jupyter-lab /gpfs/gibbs/project/support/pl543/conda_envs/ycrc_default/bin/jupyter-lab